Chemical composition, Evolutionary Origin and Nomenclature of viruses
References: Main textbook: Medical Microbiology, Jawetz, Melnick, 26th ed.,20132nd lecture of Medical Virology for 3rd Year Students
Presented by Dr. Mohammed J. M. Shallal
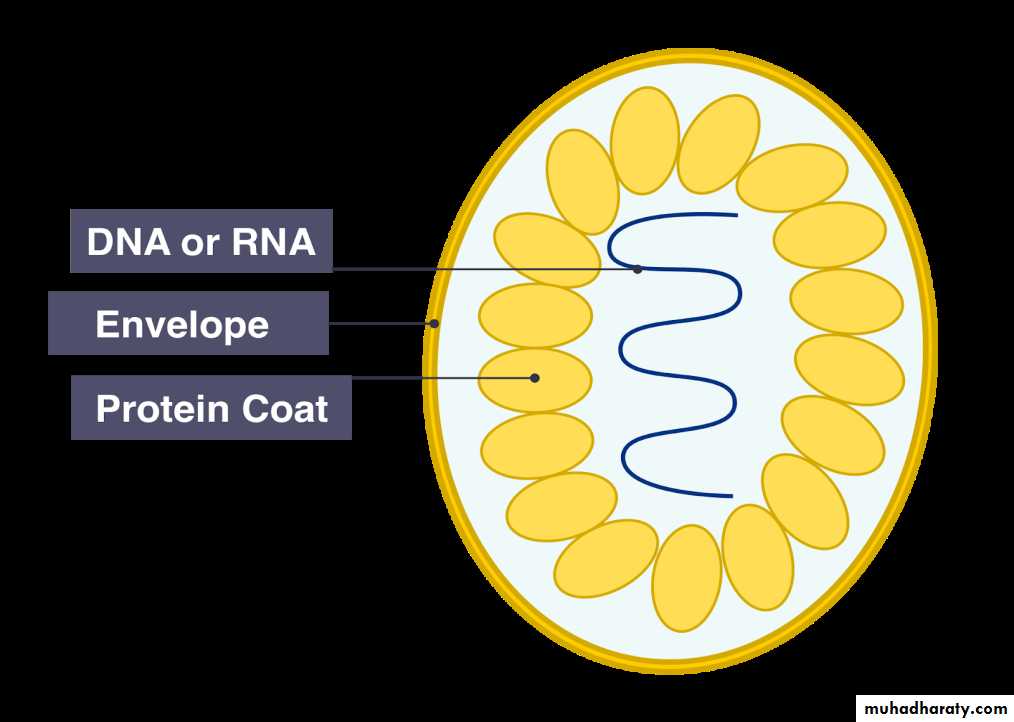
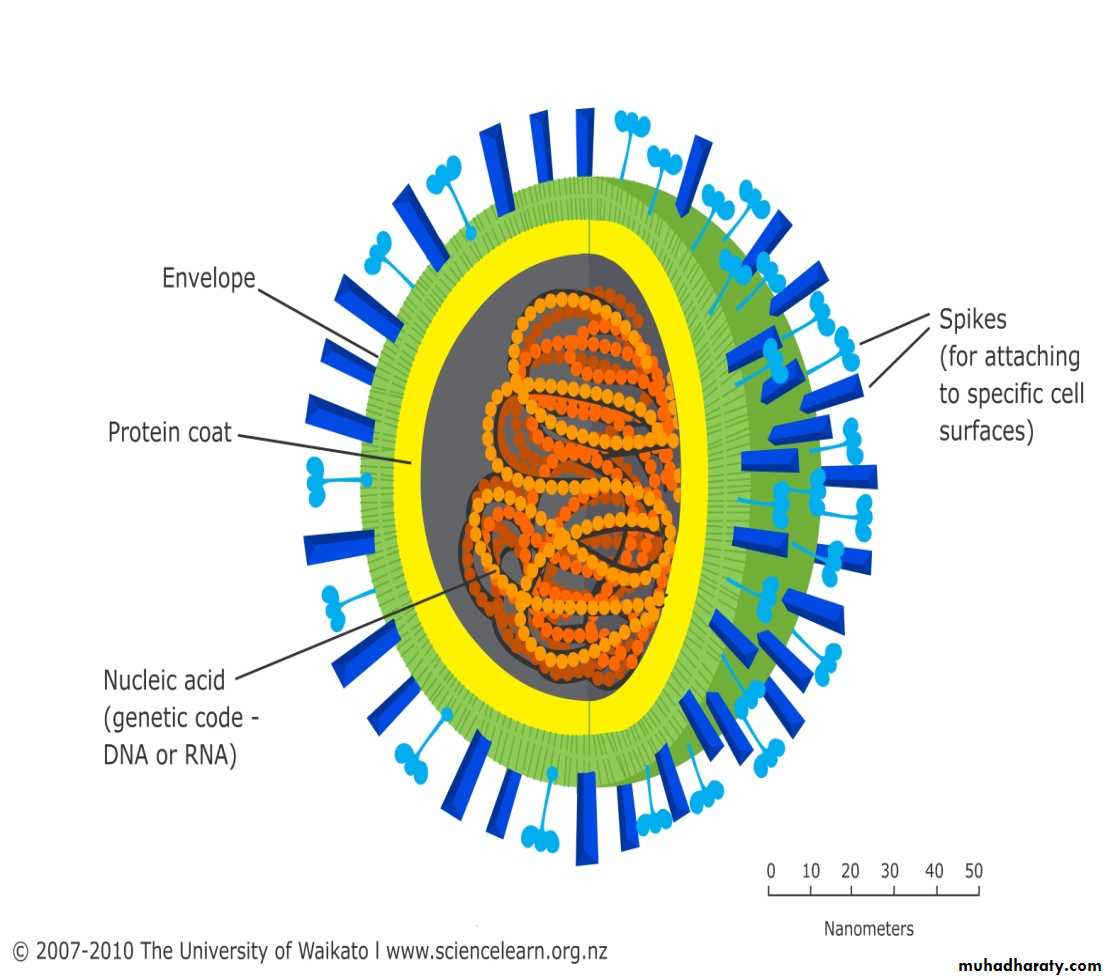
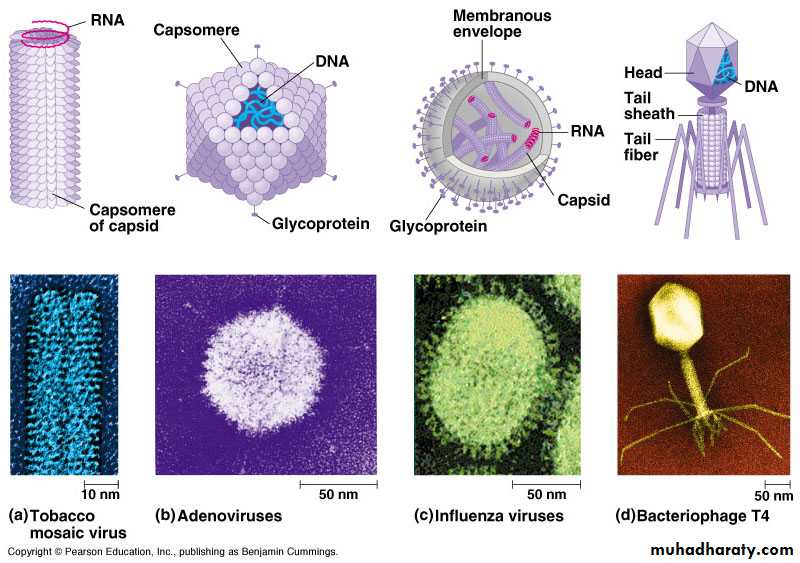
Chemical composition of virusesA) Viral protein coat:
Functions of protein shell (coat or capsid):
1- To facilitate transfer of the viral nucleic acid from one host cell to another cell2- To protect the viral genome against inactivation by nuclease enzymes.
3- Participate in the attachment of the virus particle to a susceptible cell.
4- Provide the structural symmetry of the virus particle.
5- Determine the antigenic characteristics of the virus.
6- Some surface proteins may also exhibit specific activities, eg, influenza virus hemagglutinin agglutinates red blood cells.
7- Some viruses carry enzymes (proteins) are essential for the initiation of the viral replicative cycle.
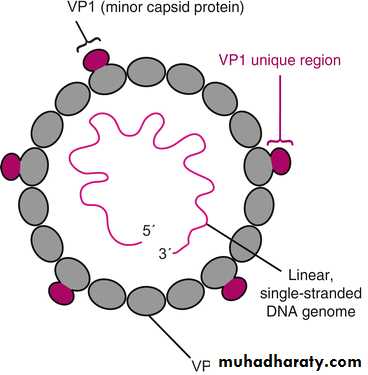
B) Viral Nucleic Acid:
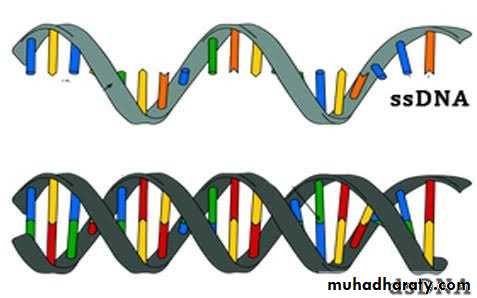
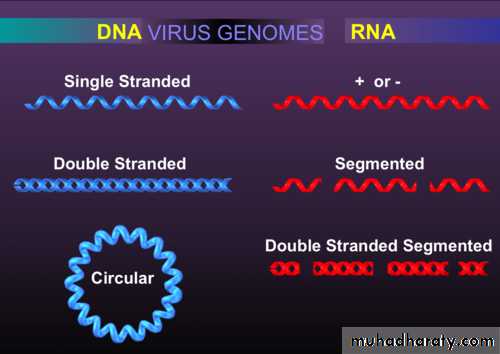
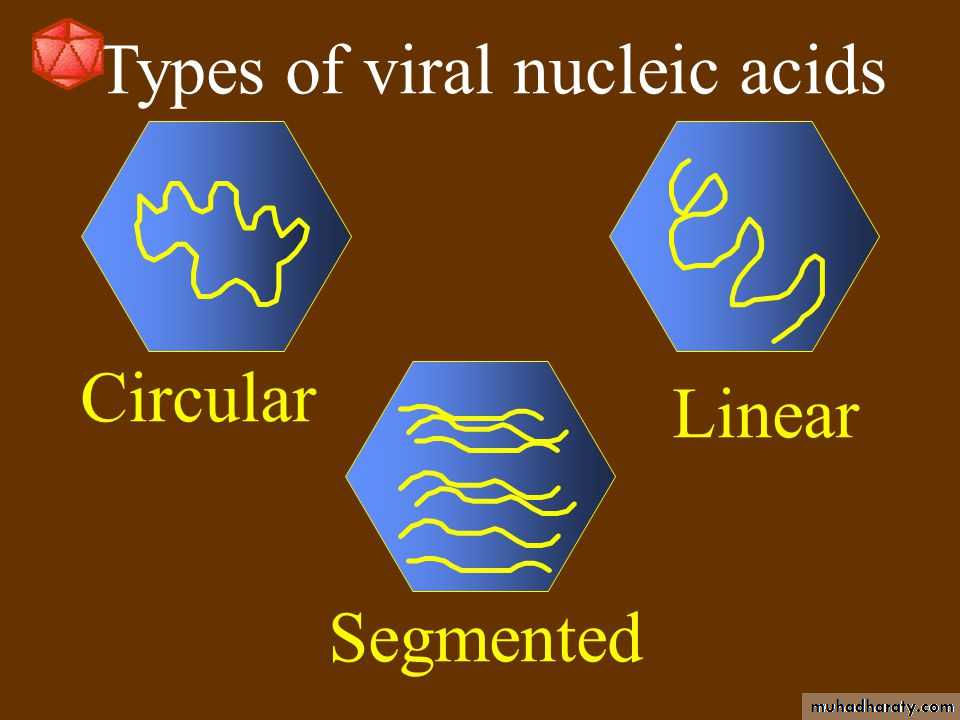
Viruses contain a single kind of nucleic acid either DNA or RNA single or double-stranded, circular or linear that encodes the genetic information necessary for replication of the virus.The size of the viral DNA genome ranges from 3.2 kbp (hepadnaviruses) to 375 kbp (poxviruses).
Size of RNA genome ranges from about 7 kb (some picornaviruses and astroviruses) to 30 kb (coronaviruses).
Viral nucleic acid may be characterized by its G + C content.
DNA viral genomes can be analyzed and compared using restriction endonucleases (enzymes that cleave DNA at specific nucleotide sequences.
Hepadnavirus
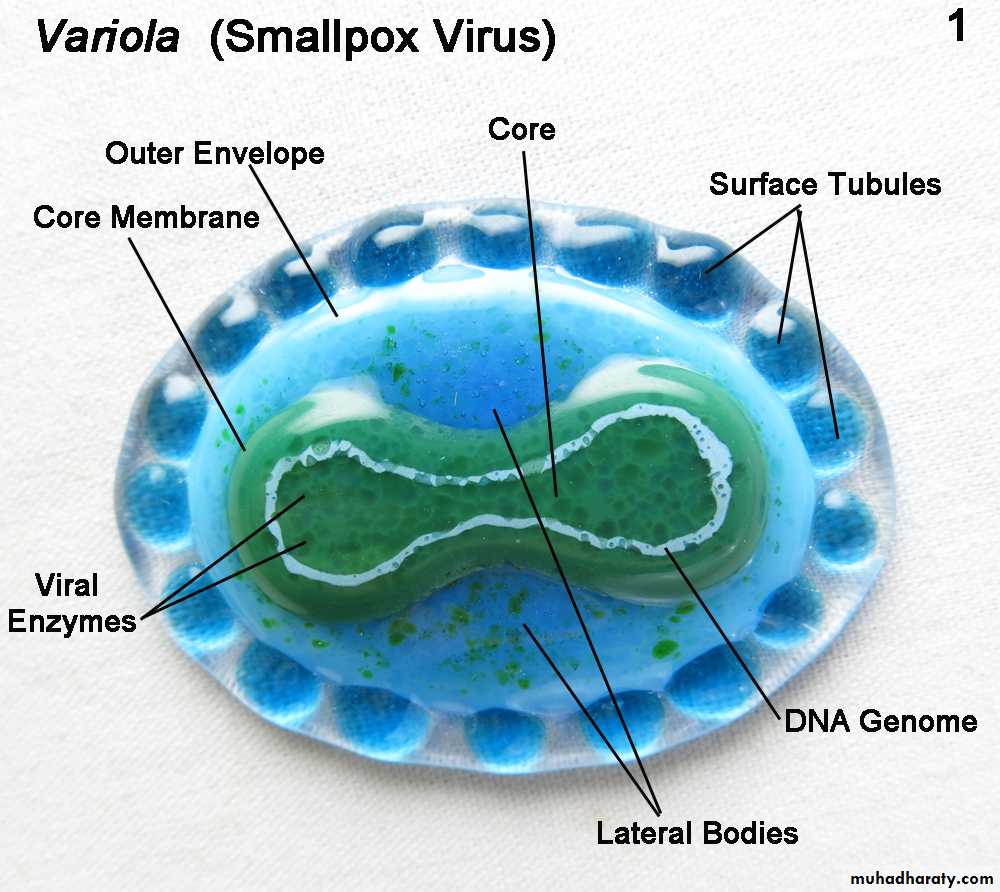
DNA genome size 3.2 kbpPoxvirus
(Small Pox- Variola)DNA genome size 375 kbp
RNA genome 7 kb (Picornavirus)
RNA genome 7 kb (Astrovirus)RNA genome 30 kb (Coronavirus)
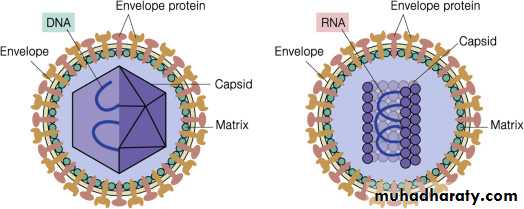
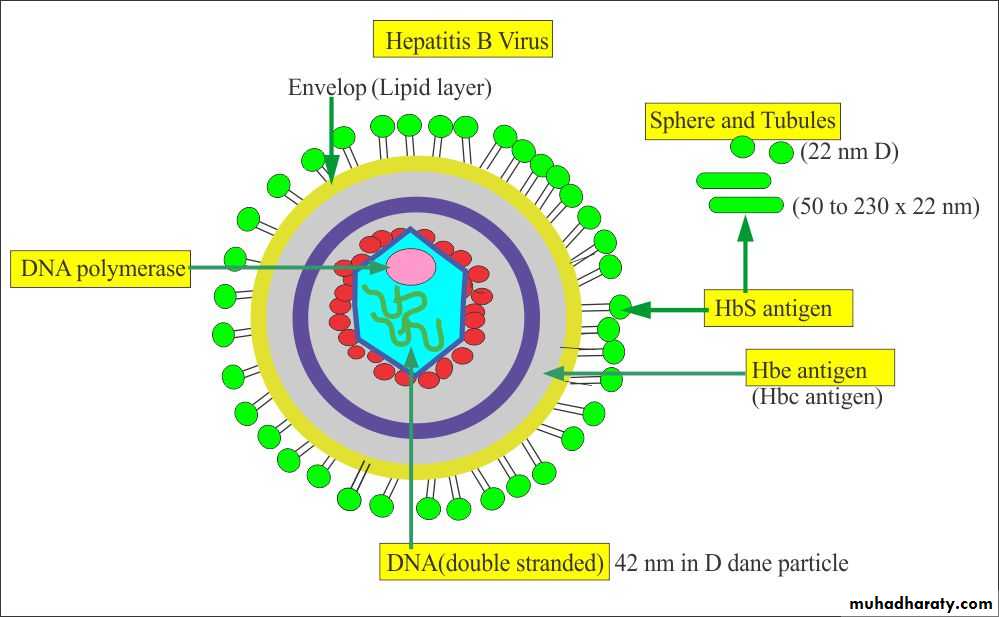
C) Viral Lipid Envelope
Some viruses contain lipid envelopes as part of their structure .The lipid is acquired when the viral nucleocapsid buds through a cellular membrane in the course of maturation. Budding occurs only at sites where virus-specific proteins have been inserted into the host cell membrane. The specific phospholipid composition of a virion envelope is determined by the specific type of cell membrane involved in the budding process. For example, herpesviruses bud through the nuclear membrane of the host cell, and the phospholipid composition of the purified virus reflects the lipids of the nuclear membrane.
Lipid-containing viruses are sensitive to effect of ether and other organic solvents indicating that disruption or loss of lipid results in loss of infectivity. On the other hand, non-lipid-containing viruses are generally resistant to Ether.
Viral envelopes also contain glycoprotein. The envelope glycoprotein is a virus-encoded. However, sugar molecules are added to viral glycoproteins often reflect the host cell in which the virus is grown and have several functions, such as:
It is the surface glycoproteins of an enveloped virus that attach the virus particle to a target cell by interacting with a cellular receptor.
They are also often involved in the membrane fusion step of infection.
The glycoproteins are also important viral antigens.
As a result of their position at the outer surface of the virion, they are frequently involved in the interaction of the virus particle with neutralizing antibody.
Extensive glycosylation of viral surface proteins may prevent effective neutralization of a virus particle by specific antibody.
Evolutionary Origin of Viruses
Three hypotheses of viral origin can be summarized as follows:
(1) Pre-cellular origin hypothesis: viruses originated before cells
(2) Escape host gene hypothesis: Viruses may be derived from DNA or RNA nucleic acid components of host cells that became able to replicate autonomously and evolve independently. Fragments of cellular genomes became infectious
(3) Regressive evolution hypothesis: cells or proto-cells evolved into virions . Viruses may be degenerate forms of intracellular parasites. There is no evidence that viruses evolved from bacteria, though other obligately intracellular organisms, eg, rickettsiae and chlamydiae, presumably did so.
Non of the above hypotheses explain the origin of virus, however the Pre-Cellular theory is most popular.
Universal System of Virus Taxonomy
A system has been established in which viruses are separated into major groupings called families on the basis of virion morphology, genome structure, and strategies of replication. Virus family names have the suffix -viridae.And each family is divided to subfamily –virinae within each subfamily, subdivisions called genera are usually based on physicochemical or serologic differences.
Criteria used to define genera vary from family to family. Genus names carry the suffix –virus.
Nomenclature of viruses
In the early days of virology, viruses were named according to common pathogenic properties: organ tropism and/or modes of transmission, and often also after their discoverers.From the early 1950s until the mid-1960s, it was popular to compose virus names by using (abbreviations derived from a few or initial letters. The name “Picornaviridae” is derived from pico (small) and RNA; the name “Reoviridae” is derived from respiratory, enteric, and orphan viruses because the agents were found in both respiratory and enteric specimens and were not related to other classified viruses; “Papovaviridae” is from papilloma, polyoma, and vacuolating agent (simian virus 40 [SV40]); “Retrovirus” is from reverse transcriptase; “Hepadnaviridae” is from the replication of the virus in hepatocytes and their DNA genomes, as seen in hepatitis B virus.
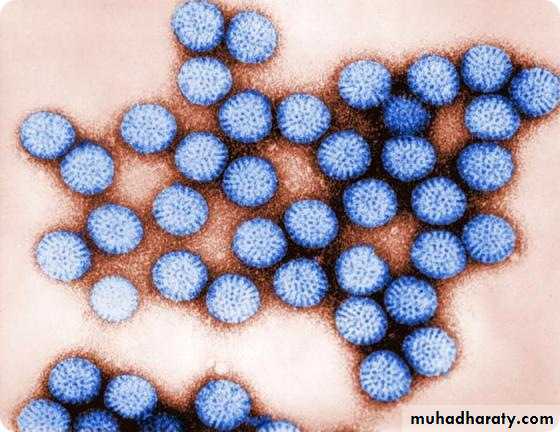
Adenoviridae (adeno, “gland”; refers to the adenoid tissue from which the viruses were first isolated); Astroviridae (astron means star); Arenaviridae (arena “sand”) describes the sandy appearance of the virion. Bunyaviridae (from Bunyamwera, the place in Africa where the type strain was isolated); Calicivirus (calix, “cup” or “goblet” from the cup-shaped depressions on the viral surfaces); Coronaviridae (corona, “crown”) describes the appearance of the peplomers protruding from the viral surface; Filoviridae (from the Latin filum, “thread” or “filament”) describes the morphology of these viruses. Herpesviridae (herpes, “creeping”) describes the nature of the lesions; Orthomyxoviridae (ortho, “true,” plus myxo “mucus,” a substance for which the viruses have an affinity; Paramyxoviridae derived from para, “closely resembling” and myxo; Parvoviridae (parvus means, “small”); Poxviridae (pock means, “pustule”); Rhabdoviridae (rhabdo, “rod” describes the shape of the viruses and Togaviridae (toga, “cloak”) refers to the tight viral envelope.
Several viruses of medical importance still remain unclassified.
Some are difficult or impossible to propagate in standard laboratory host systems and thus cannot be obtained in sufficient quantity to permit more precise characterization. Hepatitis E virus, the Norwalk virus and similar agents that cause nonbacterial gastroenteritis in humans are now assigned to the calicivirus family.
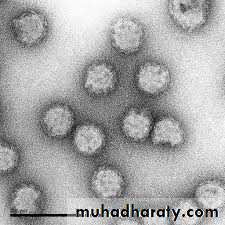
Rotavirus (derived from the Latin word “rota” meaning “wheel”)- from the characteristic wheel-like appearance of the virus when observed by electron microscopy.
Rotavirus